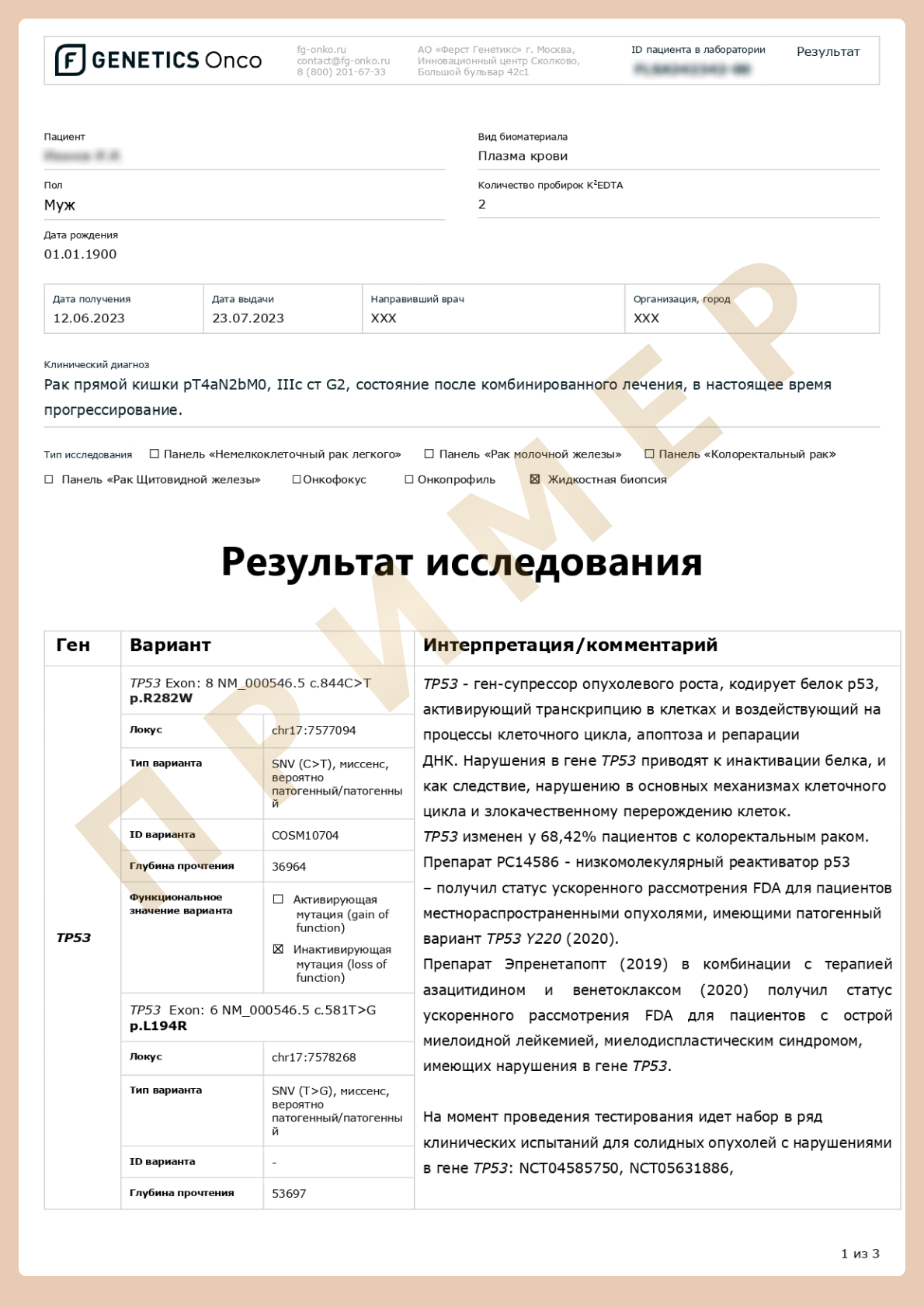
Advanced cDNA+RNA testing (liquid biopsy)
Liquid biopsy — a method for obtaining biomaterial and analysis of tumor components, primarily tumor nucleic acids in a blood sample.
Using liquid biopsy, it is possible to examine different components of biological material that can be morphologically divided into two categories:
Molecules without cells or without subcellular structure in body fluids: proteins, nucleic acids, lipids, carbohydrates, other small metabolites and metal ions.
Components with cellular or subcellular structure: single/cluster circulating tumor cells, circulating tumor-associated fibroblasts, immune cells, tumor-derived platelets, extracellular vesicles, and circulating mitochondria.
However, to date, only circulating tumor DNA and circulating tumor cells have been approved for clinical use by the US Food and Drug Administration. (FDA). [6]
Terns and abbreviations
cfDNA (cell free DNA) are short (predominant length 90-200 base pairs) DNA fragments in blood plasma, originating mainly from apoptotic and necrotic cells.
ctDNA (circulating tumor DNA) is cfDNA originating from tumor cells.
ctRNA (circulating tumor RNA) is circulating cfRNA originating from tumor cells. It is a rather unstable molecule (plasma half-life is about 15 seconds), however, when interacting with proteins, proteolipid complexes or extracellular vesicles, stability increases.
CTCs (circulating tumor cells) are tumor cells separated either from the primary tumor or from a metastatic lesion. The half-life of such a cell is from 1 to 2.4 hours [7]. The presence of CTCs in the circulation is fundamental for the development of metastasis in various types of solid tumors. However, the population of CTCs is very heterogeneous and such cells circulate in small numbers, which makes it difficult to accurately detect CTCs.
Exosomes are membrane vesicles belonging to a large class of extracellular vesicles with a diameter of 40 to 160 nm. Exosomes can mediate cellular communication under normal and pathological conditions by transporting certain molecules (nucleic acids or proteins) that reflect the composition of the cells from which they originated. Exosomes have been shown to play an important role in carcinogenesis, tumor development, metastasis, and drug resistance [8].
Tumor components in liquid biopsies and their application. Advantages and disadvantages, opportunities for use in clinical practice.
Factors affecting the release of circulating tumor nucleic acids
Circulating tumor nucleic acids (ctNA) can be passively released into the bloodstream, mainly through apoptosis and necrosis, but also through active secretion via extracellular vesicles from viable cells.
Radiation therapy
Radiation therapy induces the release of circulating tumor NA in a cell-type dependent manner. Radiation therapy causes necrosis and is considered a potential cause of necrotic release of cfDNA. However, the mechanism of cell death depends on the cell type and on the molecular aberrations present in the cells. Radiation (or chemo-) therapy can cause cellular senescence in one cell type and mitotic catastrophe in another, followed by late secondary apoptosis and necrosis.
Cellular senescence
Cellular senescence. Cellular senescence is a permanent arrest of the cell cycle caused by various internal and external stimuli such as DNA stress and damage during cytotoxic therapy. Senescence has been seen as a potential counteracting factor for cfDNA release [9].
Hypoxia
Hypoxia, a state of reduced oxygen levels in tissues, is a canonical sign of cancer that accompanies uncontrolled tumor growth. According to studies, the level of ctDNA in the blood of mice transplanted with cells of epithelial lung cancer significantly increased after exposure to intermittent hypoxia [10]. Prolonged hypoxia has been shown to negatively modulate ctDNA release [11].
Cell death
ГCell death can indirectly induce active release of circulating tumor nucleic acids through extracellular vesicles of viable cells [12].
ctNA characteristics
Half-life
The estimated half-life of ctDNA in circulation ranges from minutes to 1-2 hours. This fact indicates that cfDNA analysis reflects disease in real time. The half-life of ctDNA is related to many factors such as: encapsulation in membrane-bound vesicles, association with protein complexes, tumor type and treatment[13].
Length
The predominant cfDNA size of about 166 bp [4, 5], which corresponds to the size of mononucleosomal DNA fragments, confirms the assumption that apoptosis is the main source of cfDNA. ctDNA is usually shorter than cell-free DNA: <145 bp [14]. cfDNA of necrotic origin is longer.
Concentration
cfDNA
In general, elevated levels of cell-free DNA in the blood can be considered as an indicator of unusually high cell death associated with various pathological conditions. For example, when a tumor increases in volume, cell turnover also increases, as well as the number of apoptotic and necrotic cells. In a healthy person, cfDNA usually has a low plasma concentration with an average value of about 5 ng/mL, but varies between individuals.
ctDNA
In a malignant neoplasm, the concentration of ctDNA can vary significantly depending on tumor size, stage, localization, and other factors, with the proportion of ctDNA ranging from 0.01 to 90%.
Tumor localization and ctDNA
According to the results of a study [15],ctDNA was detected among more than 75% of patients with the following types of malignant tumors:
- advanced pancreatic cancer,
- ovarian cancer,
- colorectal cancer,
- bladder cancer,
- gastroesophageal cancer,
- breast cancer,
- melanoma,
- hepatocellular cancer,
- head and neck cancer.
However, in primary cancers of the brain, kidney, prostate, and thyroid, the detection rate of ctDNA was below 50%.
Recent studies show that the concentration of ctDNA in the blood is more correlated with characteristic features of the tumor, such as the rate of necrosis, rather than localization. For example, lung squamous cell carcinoma with a higher rate of necrosis has higher rates of ctDNA detection compared to adenocarcinomas. Also, higher levels of ctDNA were observed in triple-negative breast cancer compared to other subtypes [16],what can be explained by a higher rate of necrosis and proliferation of cells of this subtype.
A high concentration of cfDNA may be associated with a poor prognosis, but numerous studies demonstrate that the amount of cell-free DNA as a separate feature, without examining the presence of tumor mutations, is not a useful diagnostic tool [1,2, 3].
Consideration of molecular heterogeneity and tumor evolution
Molecular and cellular heterogeneity is a hallmark of cancer and one of the biggest challenges in tumor diagnosis and therapy. Unlike traditional biopsy of individual tissues, the DNA of all tumor populations, including metastases, is released into the blood; therefore, the possibilities of selecting combination therapy increase. In this regard, conducting a liquid biopsy for a limited number of genes (and especially for individual mutations in these genes) is less effective.
Clinical applications of ctDNA
ctDNA-based liquid biopsy can provide a minimally invasive, real-time assessment of tumor heterogeneity and response to treatment because it uses ctDNA released from various tumor subclones that cannot be examined with locally limited tissue biopsy.
Mutations found in ctDNA have been shown to be consistent (up to 90%) with the corresponding solid tumors [17]. Discrepancies between ctDNA and solid tumor tissue analyzes are observed mainly in patients with low levels of ctDNA [17].
Using ctDNA it is possible to track the dynamics of tumor evolution:
The blue line shows the level of ctDNA carrying the mutation found both in the primary tumor and in the blood. The yellow line shows the level of ctDNA, representing the mutation that has arisen in therapy resistance.
MRD — minimal residual disease,
WGS — whole genome sequencing,
WES — whole exome sequencing,
NGS — next generation sequencing,
LOD — limit of detection,
ddPCR — digital droplet PCR,
qPCR — quantitative PCR.
ctDNA analysis can be implemented in various clinical settings: screening and early detection of MRD, tumor assessment, treatment efficacy and relapse monitoring.
Actively released ctDNA may be of clinical importance in cancer patients at risk for dormant disseminated tumor cells (DTCs).
CtNA analysis requires an understanding of pre-analytical and analytical parameters that can affect the accuracy and reproducibility of results in a given clinical context. The main pre-analytical variables are patient-specific factors affecting the release of ctNA, sample volume, tube for sampling, storage and processing conditions. Patient-specific factors include physiological conditions (e.g., physical activity), inflammation, and both acute and chronic diseases. Therefore, the timing of plasma sampling should be carefully planned.
The amount of plasma DNA available for testing is directly proportional to the volume of plasma extracted from it, and therefore volumetric sampling must be carefully planned in advance to ensure that sufficient analyte is available to address the clinical issue.
Overview
CfNA/ctNA can be analyzed by the NGS method. For this study, the high sensitivity of the method is very important.
Our test uses unique molecular identifiers (UMI), short DNA sequences that are attached to gene-specific primers prior to amplification. Subsequently, only variants that appear in more than 80% of sequences with a particular UMI label are considered reliable. Such a system makes it possible to avoid random errors that occur during the creation of a DNA library.
The assay includes 52 genes
Advantages
- Minimal invasion
- Study of the genetic profile of tumors of hard-to-reach localization
- Display of tumor heterogeneity and systemic nature of the disease
- ctDNA can serve as a better indicator of tumor burden and heterogeneity with higher sensitivity and specificity than analysis of solid tumors
- Currently, ctDNA analysis has the highest clinical validity among biomarkers based on liquid biopsy and has more chances to being introduced into clinical practice
Limitations
- Tumor heterogeneity, evolution, and clonal hematopoiesis may partly cause ambiguity in ctDNA measurements.
Currently, there is no single ctDNA analysis that would be suitable for all purposes (e.g., early detection, MRD analysis, identification of genetic alterations and assessment of tumor genetic heterogeneity, determination of molecular mechanisms of resistance) - Risk of false-negative result with low ctNA fraction among cfNA
- Risk of false-negative result if preanalytical requirements are not taken into account

Indications for the study
- Search for markers for targeted therapy prescription
- Determination of
- metastasis predictors
- the molecular profile of hard-to-reach neoplasms
- relapse prognosis
- Monitoring of therapy
Recommendations of expert communities
ESMO (European Society of Medical Oncology) [18].
General recommendations
Validated and sensitive ctDNA testing methods can be used to detect advanced cancers and to select targeted therapy.
Primary testing by ctDNA analysis should be considered when rapid test results are needed and tissue is not available.
ctDNA testing is limited by false negative results, lower sensitivity to chimeric transcripts and gene copy number variations.
The use of ctDNA to detect minimal residual disease is not recommended due to lack of evidence for its clinical utility.
Lung cancer
The current ESMO clinical guidelines for metastatic NSCLC recommend testing all patients with non-squamous NSCLC and squamous NSCLC subtype with specific clinical characteristics (e.g., never smokers). СtDNA analysis can be performed in patients who have not previously received treatment, and it is especially advised when a significant delay in obtaining tumor tissue is expected, when invasive procedures may be risky or contraindicated, or the bone is the only area that can be biopsied.
Small, predominantly intrathoracic tumors or predominantly intracranial tumors are associated with a high false-negative rate.
A tissue RNA sequence can identify more splicing site variants such as MET exon 14 skipping and chimeric variants (e.g., ALK, RET, ROS1 or NTRK 1/2/3).
For patients with previously treated disease, the EGFR T790M resistance mutation allows treatment with osimertinib.
Breast cancer
Testing may be indicated to detect PIK3CA and ESR1 mutations in estrogen receptor (ER) positive, human epidermal growth factor receptor 2 (HER2) negative diseases, and to detect MSI as well.
Testing may be recommended to detect HER2 amplification only in situations where HER2 testing cannot be performed in conjunction with a biopsy of advanced disease because detection of HER2 amplification is suboptimal in ctDNA (assay dependent).
Mutations in ESR1 are developed subclonal in cancer, and reliable identification of the ESR1 mutation presence is more likely using fluid biopsy, tissue biopsies often give false-negative results.
Care should be taken when interpreting pathogenic variants in high penetrance cancer susceptibility genes (such as BRCA1, BRCA2, PALB2). Validated testing is required to confirm germinal or somatic origin.
Colorectal cancer
Fluid biopsy, including at least KRAS/NRAS/BRAF V600E/MSI testing for metastatic colorectal cancer without chemotherapy, is recommended when tissue testing is not possible or when a rapid therapeutic decision is required, testing for KRAS/NRAS and EGFR extracellular domain (EGFR- ECD, mutations in S492, G465, S464 and V441) is advised when restimulation with anti-EGFR monoclonal antibodies is considered.
Upper gastrointestinal tract cancer
Due to the prevalence and the availability of approved therapies, ctDNA testing on ERBB2 in gastric cancer, IDH1 and FGFR2 in cholangiocarcinoma is recommended when tissue testing is not possible or when an urgent decision is required for rapid therapeutic intervention.
General recommendations about tumor localization
Sensitivity of ctDNA analysis is reduced when central nervous system is affected by metastases and in primary brain tumors, and fluid biopsy is usually not appropriate in these patients, although it may be used if it is the only option for genetic testing. Evidence suggests that genetic testing can be done using cerebrospinal fluid analysis.
NCCN
- The NCCN supports diagnostic testing for advanced NSCLC, including molecular biomarker testing.
- The NCCN gently reminds clinicians that biomarker testing should not be used as a basis for making a histopathological diagnosis (NCCN, 2021).
- The NCCN accepts testing of cfDNA in patients when tissue testing is not available or when sufficient material for molecular analysis is not available.
- The NCCN also recognizes the need for testing in disease progression when tissue biopsy fails to detect molecular biomarkers.
How to request the Advanced cDNA+RNA testing (liquid biopsy)?
Download and fill out the informed consent form
Read the instructions for blood sampling.
It is necessary to give blood in a special tube * to stabilize cell-free DNA in any laboratory in your city. Be sure to sign the tube!
* The tube will be delivered to you by a courier to the address specified in the application, after its registration and payment for the service.
The courier will pick up the biomaterial and take it to the laboratory for free.
First Genetics Laboratory
Specialists
Years of experience in genetics, laboratory diagnostics and bioinformatics
Confidentiality
All data is strictly confidential and cannot be passed on to third parties
Time frame
Results ready in a short time
Security
Extensive control at each stage of testing
No delivery fees
Free delivery of biomaterial across Russia
Charities
Email info@f-genetics.com for information